Groups of atoms¶
MDAnalysis has a hierarchy of Atom containers that are used throughout the code.
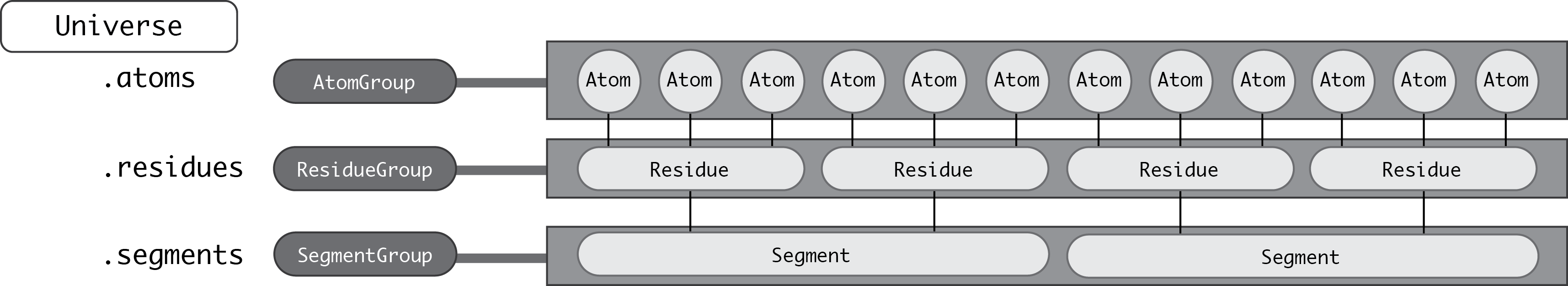
First and foremost is the AtomGroup. An AtomGroup is the primary Atom container; virtually everything can be accessed through it, as detailed in AtomGroup. This includes chemically meaningful groups of Atoms such as a Residue or a Segment.
Residues and Segments¶
A Residue is composed of Atoms, and a Segment is composed of Residues.
The corresponding container groups are ResidueGroup and SegmentGroup. These have similar properties and available methods as AtomGroup.
In [1]: import MDAnalysis as mda
In [2]: from MDAnalysis.tests.datafiles import TPR, XTC
In [3]: u = mda.Universe(TPR, XTC)
In [4]: ag = u.atoms.select_atoms('resname ARG and name CA')
In [5]: ag
Out[5]: <AtomGroup with 13 atoms>
Each of these container groups can be accessed through another. The behaviour of this differs by level. For example, the residues of the ag are the residues that the atoms of ag belong to.
In [6]: ag.residues
Out[6]: <ResidueGroup with 13 residues>
Accessing the atoms of those residues, however, returns all the atoms in the residues, not just those originally in ag.
In [7]: ag.residues.atoms
Out[7]: <AtomGroup with 312 atoms>
The same applies to segments.
In [8]: ag[:3].segments.atoms
Out[8]: <AtomGroup with 3341 atoms>
Similarly, an Atom has direct knowledge of the Residue and Segment it belongs to. Note that an Atom belongs to one Residue and the residue belongs to one Segment, but a Segment has multiple residues.
In [9]: a = u.atoms[0]
In [10]: a.residue
Out[10]: <Residue LYSH, 0>
In [11]: a.residue.segment
Out[11]: <Segment seg_0_Protein_A>
In [12]: a.residue.segment.residues
Out[12]: <ResidueGroup with 129 residues>
For information on adding custom Residues or Segments, have a look at Adding a Residue or Segment to a Universe.
Access to other classes via the AtomGroup object can be pretty powerful, but also
needs to be used with caution to avoid accessing data outside the intended selection. Therefore, we present two use
cases showing commonly used applications, for which we define Universe
on a simple extract from the PDB file:
In [9]: import MDAnalysis as mda
In [10]: import io
In [11]: pdb = io.StringIO("""
....: ATOM 414 N GLY A 402 -51.919 9.578 -14.287 1.00 68.46 N
....: ATOM 415 CA GLY A 402 -52.405 10.954 -14.168 1.00 68.41 C
....: ATOM 416 C GLY A 402 -51.821 11.946 -15.164 1.00 69.71 C
....: ATOM 417 O GLY A 402 -51.958 13.159 -14.968 1.00 69.61 O
....: ATOM 418 H GLY A 402 -52.551 8.935 -14.743 1.00 0.00 H
....: ATOM 419 HA3 GLY A 402 -52.225 11.313 -13.155 1.00 0.00 H
....: ATOM 420 HA2 GLY A 402 -53.492 10.960 -14.249 1.00 0.00 H
....: TER
....: HETATM 1929 N1 XYZ A 900 -40.275 19.399 -28.239 1.00 0.00 N1+
....: TER
....: ATOM 1029 N ALA B 122 -25.408 19.612 -13.814 1.00 37.52 N
....: ATOM 1030 CA ALA B 122 -26.529 20.537 -14.038 1.00 37.70 C
....: ATOM 1031 C ALA B 122 -26.386 21.914 -13.374 1.00 45.35 C
....: ATOM 1032 O ALA B 122 -26.885 22.904 -13.918 1.00 48.34 O
....: ATOM 1033 CB ALA B 122 -27.835 19.889 -13.613 1.00 37.94 C
....: ATOM 1034 H ALA B 122 -25.636 18.727 -13.385 1.00 0.00 H
....: ATOM 1035 HA ALA B 122 -26.592 20.707 -15.113 1.00 0.00 H
....: ATOM 1036 HB1 ALA B 122 -28.658 20.583 -13.783 1.00 0.00 H
....: ATOM 1037 HB2 ALA B 122 -27.998 18.983 -14.196 1.00 0.00 H
....: ATOM 1038 HB3 ALA B 122 -27.788 19.635 -12.554 1.00 0.00 H
....: ATOM 1039 N GLY B 123 -25.713 21.969 -12.223 1.00 41.18 N
....: ATOM 1040 CA GLY B 123 -25.550 23.204 -11.460 1.00 41.40 C
....: ATOM 1041 C GLY B 123 -24.309 24.018 -11.745 1.00 45.74 C
....: ATOM 1042 O GLY B 123 -24.349 25.234 -11.601 1.00 46.81 O
....: ATOM 1043 H GLY B 123 -25.290 21.133 -11.845 1.00 0.00 H
....: ATOM 1044 HA3 GLY B 123 -25.593 22.976 -10.395 1.00 0.00 H
....: ATOM 1045 HA2 GLY B 123 -26.430 23.831 -11.600 1.00 0.00 H
....: TER
....: """)
....:
In [12]: u = mda.Universe(pdb, format="PDB")
Use case: Sequence of residues by segment¶
In order to select only ATOM record types and get a list of residues by segment, one needs to call:
In [13]: residues_by_seg = list()
In [14]: for seg in u.segments:
....: p_seg = seg.atoms.select_atoms("record_type ATOM")
....: residues_by_seg.append(p_seg.residues)
....:
Residue names can be extracted using Python’s list comprehensions. As required, HETATM record type lines are not considered:
In [15]: [rg.resnames for rg in residues_by_seg]
Out[15]: [array(['GLY'], dtype=object), array(['ALA', 'GLY'], dtype=object)]
Note that accessing residues by first selecting the segments of an AtomGroup returns
all the residues in that segment for both the ATOM and HETATM record types (no memory of the original selection).
The meaning of this is: “give me all residue names from segments in which there is at least one of the selected atoms”.
In [16]: selected_atoms = u.select_atoms("record_type ATOM")
In [17]: all_residues = selected_atoms.segments.residues
In [18]: all_residues.resnames
Out[18]: array(['GLY', 'XYZ', 'ALA', 'GLY'], dtype=object)
Use case: Atoms list grouped by residues¶
In order to list all the heavy protein backbone and sidechain atoms in every residue, one needs to call:
In [19]: atoms_in_residues = list()
In [20]: for seg in u.segments:
....: p_seg = seg.atoms.select_atoms("record_type ATOM and not name H*")
....: for p_res in p_seg.residues:
....: atoms_in_residues.append(p_seg.select_atoms(f"resid {p_res.resid} and resname {p_res.resname}"))
....:
Atom names can be extracted using Python’s list comprehensions. As required, HETATM record type lines and hydrogen atoms are not considered:
In [21]: [ag.names for ag in atoms_in_residues]
Out[21]:
[array(['N', 'CA', 'C', 'O'], dtype=object),
array(['N', 'CA', 'C', 'O', 'CB'], dtype=object),
array(['N', 'CA', 'C', 'O'], dtype=object)]
The Python syntax can be further simplified by using split() function:
In [22]: rds = u.select_atoms("record_type ATOM and not name H*").split("residue")
In [23]: [ag.names for ag in rds]
Out[23]:
[array(['N', 'CA', 'C', 'O'], dtype=object),
array(['N', 'CA', 'C', 'O', 'CB'], dtype=object),
array(['N', 'CA', 'C', 'O'], dtype=object)]
Note that accessing atoms by first selecting the residues of an AtomGroup also
returns hydrogen atoms (no memory of the original selection). The meaning of this is “give me all atom names from
residues in which there is at least one of the selected atoms”. However, it doesn’t contain a nitrogen atom from XYZ
residue as no atoms from this residue were in the AtomGroup.
In [24]: all_atoms_in_residues = list()
In [25]: for seg in u.segments:
....: p_seg = seg.atoms.select_atoms("record_type ATOM and not name H*")
....: all_atoms_in_residues.append(p_seg.residues)
....:
In [26]: [atom.names for atom in all_atoms_in_residues]
Out[26]:
[[array(['N', 'CA', 'C', 'O', 'H', 'HA3', 'HA2'], dtype=object)],
[array(['N', 'CA', 'C', 'O', 'CB', 'H', 'HA', 'HB1', 'HB2', 'HB3'],
dtype=object),
array(['N', 'CA', 'C', 'O', 'H', 'HA3', 'HA2'], dtype=object)]]
Fragments¶
Certain analysis methods in MDAnalysis also make use of additional ways to group atoms. A key concept is a fragment. A fragment is what is typically considered a molecule: an AtomGroup where any atom is reachable from any other atom in the AtomGroup by traversing bonds, and none of its atoms is bonded to any atoms outside the AtomGroup. (A ‘molecule’ in MDAnalysis methods refers to a GROMACS-specific concept). The fragments of a Universe are determined by MDAnalysis as a derived quantity. They can only be determined if bond information is available.
The fragments of an AtomGroup are accessible via the fragments property. Below is a Universe from a GROMACS TPR file of lysozyme (PDB ID: 2LYZ) with 101 water molecules. While it has 230 residues, there are only 102 fragments: 1 protein and 101 water fragments.
In [12]: len(u.residues)
Out[12]: 230
In [13]: len(u.atoms.fragments)
Out[13]: 102
See Topology objects for more on bonds and which file formats give MDAnalysis bond information.
You can also look at which fragment a particular Atom belongs to:
In [27]: u.atoms[0].fragment # first atom of lysozyme
---------------------------------------------------------------------------
NoDataError Traceback (most recent call last)
<ipython-input-27-f5f6a0558be2> in <module>
----> 1 u.atoms[0].fragment # first atom of lysozyme
/opt/anaconda3/envs/mda-user-guide/lib/python3.8/site-packages/MDAnalysis/core/groups.py in __getattr__(self, attr)
4145 elif attr == 'position':
4146 raise NoDataError('This Universe has no coordinates')
-> 4147 return super(Atom, self).__getattr__(attr)
4148
4149 @property
/opt/anaconda3/envs/mda-user-guide/lib/python3.8/site-packages/MDAnalysis/core/groups.py in __getattr__(self, attr)
4025 raise NoDataError(err.format(singular=cls.singular))
4026 else:
-> 4027 return super(ComponentBase, self).__getattr__(attr)
4028
4029 def __lt__(self, other):
/opt/anaconda3/envs/mda-user-guide/lib/python3.8/site-packages/MDAnalysis/core/groups.py in __getattr__(self, attr)
374 err = ('{selfcls}.{attrname} not available; '
375 'this requires {topattr}')
--> 376 raise NoDataError(err.format(selfcls=selfcls,
377 attrname=attrname,
378 topattr=topattr))
NoDataError: Atom.fragment not available; this requires bonds
and see which fragments are associated with atoms in a smaller AtomGroup:
In [28]: u.atoms[1959:1961].fragments
---------------------------------------------------------------------------
NoDataError Traceback (most recent call last)
<ipython-input-28-9bf24618be6c> in <module>
----> 1 u.atoms[1959:1961].fragments
/opt/anaconda3/envs/mda-user-guide/lib/python3.8/site-packages/MDAnalysis/core/groups.py in __getattr__(self, attr)
2535 elif attr == 'positions':
2536 raise NoDataError('This Universe has no coordinates')
-> 2537 return super(AtomGroup, self).__getattr__(attr)
2538
2539 @property
/opt/anaconda3/envs/mda-user-guide/lib/python3.8/site-packages/MDAnalysis/core/groups.py in __getattr__(self, attr)
609 raise NoDataError(err.format(singular=cls.singular))
610 else:
--> 611 return super(GroupBase, self).__getattr__(attr)
612
613 def __repr__(self):
/opt/anaconda3/envs/mda-user-guide/lib/python3.8/site-packages/MDAnalysis/core/groups.py in __getattr__(self, attr)
374 err = ('{selfcls}.{attrname} not available; '
375 'this requires {topattr}')
--> 376 raise NoDataError(err.format(selfcls=selfcls,
377 attrname=attrname,
378 topattr=topattr))
NoDataError: AtomGroup.fragments not available; this requires bonds
Note
AtomGroup.fragments returns a tuple of fragments with at least one Atom in the AtomGroup, not a tuple of fragments where all Atoms are in the AtomGroup.